Gut Microbiota in Health and Disease

Ms Wilson’s article reviews the influence of the environment on the gut microbiome and discusses potential dietary avenues to improve it.
As new evidence and better analytical techniques emerge, more information is becoming available about our gut bacteria. It is becoming clear that the type and relative amount of bacteria present in our gut plays an important role in both health and disease. Biotech companies are investing more in technologies to target this ‘microbiome’ as a potential moderator of our gut health and our innate immune system. The increase in incidence of immune mediated diseases and neurological disorders cannot be explained by shifts in human genetics. [1] Dysbiosis and loss of diversity is something that is now being commonly traced to these diseases as we look to our ‘other genome’ for clues. What has become abundantly clear from the work thus far conducted in metagenomics is that greater bacterial diversity or ‘gene richness’ is strongly associated with better health.
INFO Key definitions
- Microbiome: the collective name for the gut microorganisms.
- Dysbiosis: one or more potentially harmful microbial organisms predominating the gut microbial population. [3]
- Metagenomics: the field of study which compares entire genomes.
- Phyla: a taxonomic term for dividing organisms into groups with others of similar properties.
- Enterotype: a term to divide humans into groups based on the gut bacteria present.
- Transcriptome: the genes which have been expressed as proteins i.e. the active part of the genome.
What species makes up the Microbiome?
Although great inter-individual variation exists at species level – most people have around 160 species from a possible 1000 – the phyla represented in the microbiome are quite narrow. Qin et al (2010) have identified a core set of bacteria present within all people. Three main enterotypes have been configured for the human microbiome. [2] The marker genera for which microbiome a subject belongs to are the Bacteroides, Prevotella and Ruminococcus (the latter group is further associated with the prescence of the Methanobrevibacter). [1]
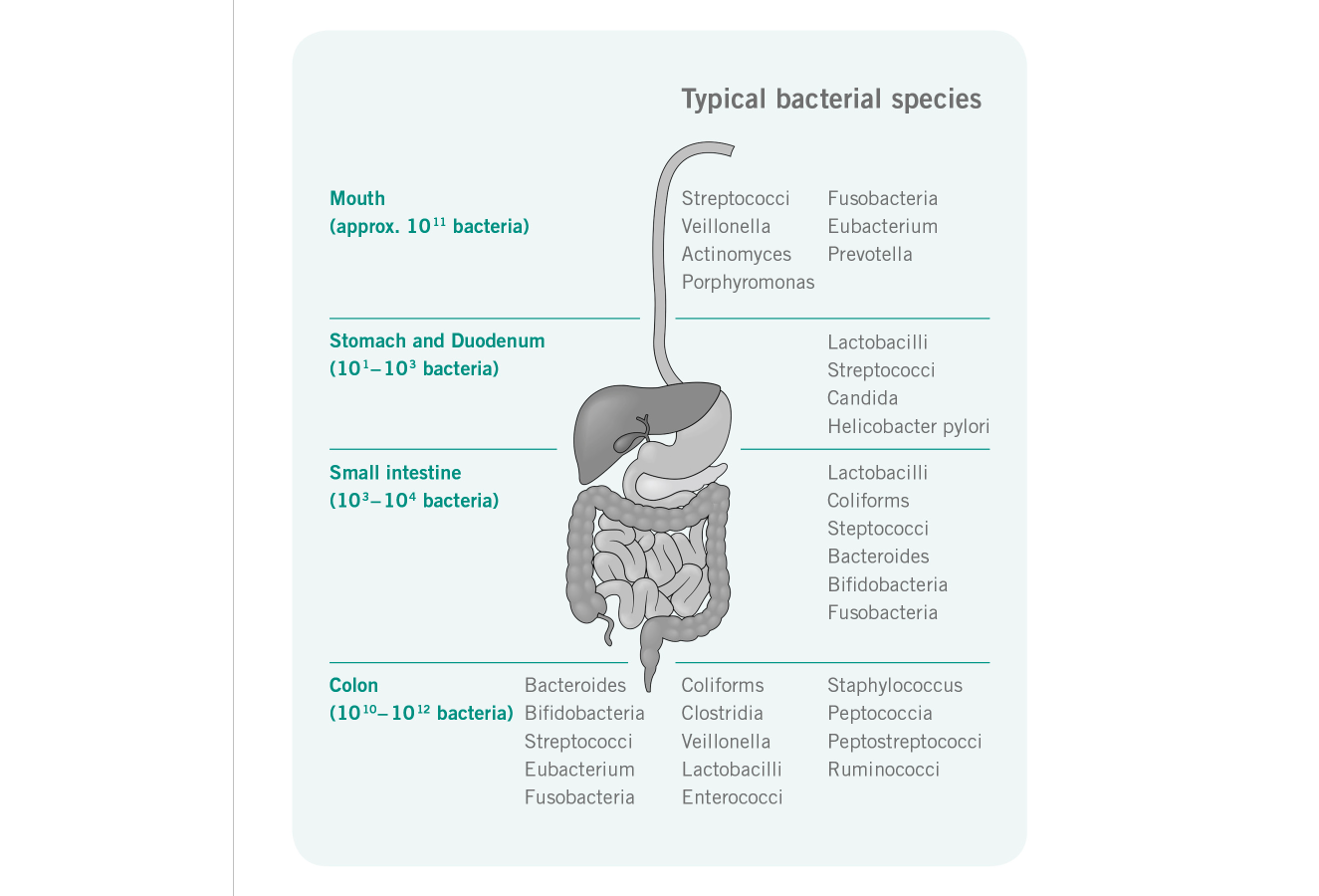
Function
The functions of the gut microbiota are still being fully elucidated but some key aspects are: immune signaling and modulation; production of nervous system messengers; production of essential vitamins; regulation of fat metabolism; production of short chain fatty acids (SCFA) specifically butyrate, and branched chain fatty acids. Depending on the substrate fermented, other products of the microbiome include hydrogen, carbon-dioxide and methane gasses, ammonia, amines and phenolic compounds. [3]
The symbiosis between humans and the gut microbiota is becoming ever more apparent. The term ‘superorganism’ has been coined as the human body starts to be considered more a conglomerate organism of our own transcriptome and far greater transcriptome of the gut microbiota. The genes encoded by our gut bacteria outstrip our own by greater than 100-fold. [4] It is therefore not surprising that much focus of disease cause, prevention and cure is now being placed on this ‘other genome’.
The enteric nervous system (ENS) is sometimes referred to in literature as the ‘second brain’. This is due to it consisting of over 200 million neurons. [5] The ENS sends signals from the gut to the brain via endocrine, neuronal and immune afferent signaling. [5] In addition, the gut associated lymphoid tissue (GALT), which regularly samples and responds to signals from within the intestinal lumen, is considered the bodies major defensive organ against infection. [5]
The combination of interactions between the ENS the microbiome and the GALT has great potential to effect changes to physical, immunological and emotional wellbeing.
The symbiosis between humans and the gut microbiota is becoming ever more apparent. The term ‘superorganism’ has been coined as the human body starts to be considered more a conglomerate organism of our own transcriptome and far greater transcriptome of the gut microbiota. The genes encoded by our gut bacteria outstrip our own by greater than 100-fold. [4] It is therefore not surprising that much focus of disease cause, prevention and cure is now being placed on this ‘other genome’.
The enteric nervous system (ENS) is sometimes referred to in literature as the ‘second brain’. This is due to it consisting of over 200 million neurons. [5] The ENS sends signals from the gut to the brain via endocrine, neuronal and immune afferent signaling. [5] In addition, the gut associated lymphoid tissue (GALT), which regularly samples and responds to signals from within the intestinal lumen, is considered the bodies major defensive organ against infection. [5]
The combination of interactions between the ENS the microbiome and the GALT has great potential to effect changes to physical, immunological and emotional wellbeing.
What shapes the Microbiome?
The microbiome develops from birth. Route of delivery and early feeding affects the initial development of the microbiome. Weaning and childhood environment (rural or urban) likely has an impact on the development of the mature microbiome. Studies of isolated population groups in Africa show divergent and unique bacterial colonization from that of a Western cohort. These findings indicate that the environment is a strong driving force for colonization. [6] Twin studies have also revealed that at least with some taxa there is a definite genetic influence on species abundance. [7] The spouses of identical twins also showed positive correlations which adds to the concept that both nature and nurture can affect gene richness of the gut microbiota. [8]
In the elderly the microbiome changes again, although it is not clear why this occurs. A reduction in butyrate producing bacteria is seen within the elderly population and a reduction in gene richness of the microbiome. Elderly living within the community maintain greater gene richness, thought to be an artefact of a more varied diet, than elderly in long term care. [9]
In the elderly the microbiome changes again, although it is not clear why this occurs. A reduction in butyrate producing bacteria is seen within the elderly population and a reduction in gene richness of the microbiome. Elderly living within the community maintain greater gene richness, thought to be an artefact of a more varied diet, than elderly in long term care. [9]
How do changes affect us?
Dysbiosis is associated with several disease states. [3] A recent review was published on diseases and the associated alterations in bacterial populations. [4] New technologies have allowed a type of bacterial fingerprint to be developed for certain diseases which may provide both a powerful and non-invasive diagnostic tool and a potential target for therapy in treating these conditions.
In obesity studies, subjects with reduced gene richness showed greater overall adiposity, insulin resistance and dyslipidaemia and a more pronounced inflammatory phenotype. [10] Those with lower gene richness also gained more weight over time. Goodrich et al (2014) provide insight into the heritability of an obesogenic microbiome and the potential influence of methanogens and Christensenella spp. on metabolic disorders.
Studies in infants with genetic disposition to celiac disease (CD) show a reduction in Actinobacteria (which includes Bifidobacteria) and an increase in Firmicutes and Proteobacteria spp. [11] This said, a definitive link between microbial alterations and development of CD disease has not been shown. [12]
Dysbiosis has been identified in microbiome sequencing in irritable bowel syndrome (IBS). [13] Further studies have identified distinct differences in gut bacteria in different IBS subtypes in both luminal and mucosal populations. [14-16] A recent pilot study in pediatrics’ has identified that the microbiome may be indicative of the likelihood of the low FODMAP diet being effective in symptom alleviation. [17]
Qin et al (2012) identified antagonistic behavior between beneficial and harmful bacteria in type 2 diabetes. A decline in butyrate producing bacteria may be an indicator of increased risk of developing obesity related co-morbidities. [18]
In obesity studies, subjects with reduced gene richness showed greater overall adiposity, insulin resistance and dyslipidaemia and a more pronounced inflammatory phenotype. [10] Those with lower gene richness also gained more weight over time. Goodrich et al (2014) provide insight into the heritability of an obesogenic microbiome and the potential influence of methanogens and Christensenella spp. on metabolic disorders.
Studies in infants with genetic disposition to celiac disease (CD) show a reduction in Actinobacteria (which includes Bifidobacteria) and an increase in Firmicutes and Proteobacteria spp. [11] This said, a definitive link between microbial alterations and development of CD disease has not been shown. [12]
Dysbiosis has been identified in microbiome sequencing in irritable bowel syndrome (IBS). [13] Further studies have identified distinct differences in gut bacteria in different IBS subtypes in both luminal and mucosal populations. [14-16] A recent pilot study in pediatrics’ has identified that the microbiome may be indicative of the likelihood of the low FODMAP diet being effective in symptom alleviation. [17]
Qin et al (2012) identified antagonistic behavior between beneficial and harmful bacteria in type 2 diabetes. A decline in butyrate producing bacteria may be an indicator of increased risk of developing obesity related co-morbidities. [18]
How can we improve the Microbiome?
Dietary studies have shown the power of dietary manipulation in altering the microbiome [19] and this is one area with great potential. Feeding the host and feeding the microbiota is the obvious way in which manipulation of the microbiota may come about. A high fat-high protein diet has been linked to the Bacteroides enterotype, and a carbohydrate rich diet corresponds to the Prevotella enterotype. [20] Short term dietary changes (~10 days) were shown to change the composition of the microbiome but not significantly affect the identity of the enterotype. Faecalibaterium prausnitzii, Bifidobacterium and Clostridium cluster XIVa have all been shown to be elevated by high fiber dietary supplements and these are three groups which are generally associated with better health. [1,21]
Other studies have definitively shown that prebiotics and probiotics to varying degrees are useful tools in promoting beneficial bifidobacteria and lactobacilli. Several studies have shown mechanisms by which different species of Lactobacilli and Bifidobacteria not only confer beneficial effects on the host but also inhibit attachment and activity of invading enteropathogens. [4] It may be that in the future more strains of bacteria (e.g. Akkermansia mucinophila and Christensenella minuta) will be targeted by both prebiotic and probiotic supplementation. [4]
Fecal microbiota transplantation is another technique for rapid correction of a disordered microbiome. Trials for intractable C.diff infected patients have shown promising results thus far. Lean donor fecal microbiota transplantation showed an improvement in insulin resistance in patients with metabolic syndrome [22] – lending support to the growing concept that dysbiosis plays an important role in the development of obesity related disorders. Antimicrobial strategies for modulation of the microbiome may have therapeutic potential in the future however current knowledge of the efficacy in this area is based on mice models and therefore not presently a recommended strategy. [4]
A varied diet including all food groups should be utilized to provide alternating substrate and reduce the likelihood of unfavorable species becoming dominant within the gut. For perturbations in the microbiome brought on by antibiotics or a bout of gastroenteritis the use of prebiotics and probiotics are likely to be useful and are considered to be safe for general use. Stool diagnostics capable of detecting a decrease in diversity may be a useful tool for early disease prediction and preventative measures.
Other studies have definitively shown that prebiotics and probiotics to varying degrees are useful tools in promoting beneficial bifidobacteria and lactobacilli. Several studies have shown mechanisms by which different species of Lactobacilli and Bifidobacteria not only confer beneficial effects on the host but also inhibit attachment and activity of invading enteropathogens. [4] It may be that in the future more strains of bacteria (e.g. Akkermansia mucinophila and Christensenella minuta) will be targeted by both prebiotic and probiotic supplementation. [4]
Fecal microbiota transplantation is another technique for rapid correction of a disordered microbiome. Trials for intractable C.diff infected patients have shown promising results thus far. Lean donor fecal microbiota transplantation showed an improvement in insulin resistance in patients with metabolic syndrome [22] – lending support to the growing concept that dysbiosis plays an important role in the development of obesity related disorders. Antimicrobial strategies for modulation of the microbiome may have therapeutic potential in the future however current knowledge of the efficacy in this area is based on mice models and therefore not presently a recommended strategy. [4]
A varied diet including all food groups should be utilized to provide alternating substrate and reduce the likelihood of unfavorable species becoming dominant within the gut. For perturbations in the microbiome brought on by antibiotics or a bout of gastroenteritis the use of prebiotics and probiotics are likely to be useful and are considered to be safe for general use. Stool diagnostics capable of detecting a decrease in diversity may be a useful tool for early disease prediction and preventative measures.
Author
BRIDGETTE WILSON BSC, MSC, PGDIP, RD.
Research Dietitian, King’s College London. Bridgette is a Doctoral Student at King’s College London and a registered Dietitian. Bridgette Completed a Bachelor of Biological Sciences and a Masters in Molecular Biology before training as a Dietitian. Following on from working in the NHS Bridgette returned to a research role to focus on the area of Gastroenterology. Bridgette currently works under the team of Professor Kevin Whelan and Dr Miranda Lomer at King’s College London and is undertaking research into dietary interventions for irritable bowel syndrome.
Research Dietitian, King’s College London. Bridgette is a Doctoral Student at King’s College London and a registered Dietitian. Bridgette Completed a Bachelor of Biological Sciences and a Masters in Molecular Biology before training as a Dietitian. Following on from working in the NHS Bridgette returned to a research role to focus on the area of Gastroenterology. Bridgette currently works under the team of Professor Kevin Whelan and Dr Miranda Lomer at King’s College London and is undertaking research into dietary interventions for irritable bowel syndrome.
References
- Blottiere, H.M., et al., Human intestinal metagenomics: state of the art and future. Curr Opin Microbiol, 2013. 16(3): p. 232-9.
- Qin, J., et al., A human gut microbial gene catalogue established by metagenomic sequencing. Nature, 2010. 464(7285): p. 59-65.
- Roberfroid, M., et al., Prebiotic effects: metabolic and health benefits. British Journal of Nutrition, 2010. 104(S2): p. S1-S63.
- Walsh, C.J., et al., Beneficial modulation of the gut microbiota. FEBS letters, 2014. 588(22): p. 4120-4130.
- Al Omran, Y. and Q. Aziz, The brain-gut axis in health and disease, in Microbial Endocrinology: The Microbiota-Gut-Brain Axis in Health and Disease. 2014, Springer. p. 135-153.
- De Filippo, C., et al., Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proceedings of the National Academy of Sciences, 2010. 107(33): p. 14691-14696.
- Goodrich, J.K., et al., Human genetics shape the gut microbiome. Cell, 2014. 159(4): p. 789-799.
- Nelson, K.E., et al., Metagenomics of the human body. 2011: Springer.
- Claesson, M.J., et al., Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proceedings of the National Academy of Sciences, 2011. 108(Supplement 1): p. 4586-4591.
- Le Chatelier, E., et al., Richness of human gut microbiome correlates with metabolic markers. Nature, 2013. 500(7464): p. 541-546.
- Olivares, M., et al., The HLA-DQ2 genotype selects for early intestinal microbiota composition in infants at high risk of developing coeliac disease. Gut, 2014: p. gutjnl-2014-306931.
- McLean, M.H., et al., Does the microbiota play a role in the pathogenesis of autoimmune diseases? Gut, 2014: p. gutjnl-2014-308514.
- Rajilic-Stojanovic, M., et al., Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology, 2011. 141(5): p. 1792-801.
- Saulnier, D.M., et al., The intestinal microbiome, probiotics and prebiotics in neurogastroenterology. Gut Microbes, 2013. 4(1): p. 17-27.
- Parkes, G.C., et al., Distinct microbial populations exist in the mucosa-associated microbiota of subgroups of irritable bowel syndrome. Neurogastroenterology & Motility, 2012. 24(1): p. 31-39.
- Sundin, J., et al., Altered faecal and mucosal microbial composition in post-infectious irritable bowel syndrome patients correlates with mucosal lymphocyte phenotypes and psychological distress. Aliment Pharmacol Ther, 2015. 41(4): p. 342-51.
- Chumpitazi, B.P., et al., Gut microbiota influences low fermentable substrate diet efficacy in children with irritable bowel syndrome. Gut microbes, 2014. 5(2): p. 165-175.
- Qin, J., et al., A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature, 2012. 490(7418): p. 55-60.
- Cotillard, A., et al., Dietary intervention impact on gut microbial gene richness. Nature, 2013. 500(7464): p. 585-8.
- Wu, G.D., et al., Linking long-term dietary patterns with gut microbial enterotypes. Science, 2011. 334(6052): p. 105-108.
- Shen, Q., L. Zhao, and K.M. Tuohy, High-level dietary fibre up-regulates colonic fermentation and relative abundance of saccharolytic bacteria within the human faecal microbiota in vitro. European journal of nutrition, 2012. 51(6): p. 693-705.
- Vrieze, A., et al., Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology, 2012. 143(4): p. 913-6.e7.
www.drschaer-institute.com
